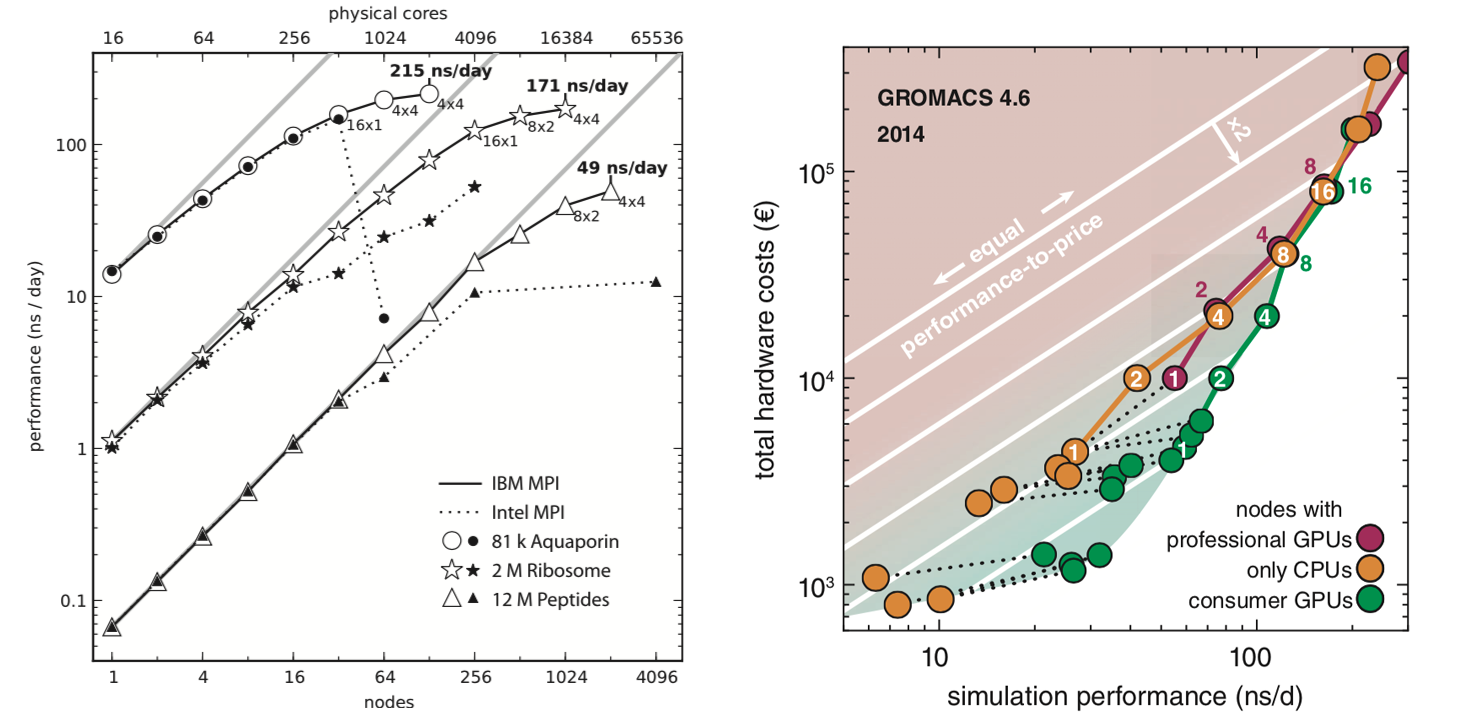
Best bang for your buck: GPU nodes for GROMACS biomolecular simulations - Kutzner - 2015 - Journal of Computational Chemistry - Wiley Online Library
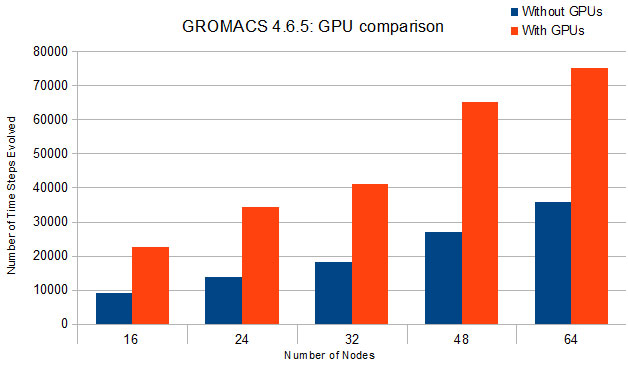
More Bang for Your Buck: Improved use of GPU Nodes for GROMACS 2018 arXiv:1903.05918v2 [cs.DC] 13 Jun 2019

GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers - ScienceDirect